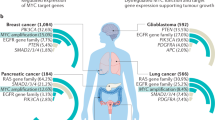
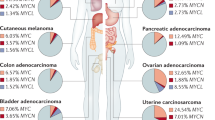
Abstract
Is the oncogene MYC upregulated or hyperactive? In the majority of human cancers, finding agents that target c-MYC has proved difficult. Here we report specific bacterial effector molecules that inhibit cellular MYC (c-MYC) in human cells. We show that uropathogenic Escherichia coli (UPEC) degrade the c-MYC protein and attenuate MYC expression in both human cells and animal tissues. c-MYC protein was rapidly degraded by both cell-free bacterial lysates and the purified bacterial protease Lon. In mice, intravesical or peroral delivery of Lon protease delayed tumor progression and increased survival in MYC-dependent bladder and colon cancer models, respectively. These results suggest that bacteria have evolved strategies to control c-MYC tissue levels in the host and that the Lon protease shows promise for therapeutic targeting of c-MYC in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The microarray data for this study have been deposited in NCBI’s Gene Expression Omnibus repository under accession number GSE162253. All other data that support the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Meyer, N. & Penn, L. Z. MYC – TIMELINE reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008).
Nilsson, J. A. & Cleveland, J. L. Myc pathways provoking cell suicide and cancer. Oncogene 22, 9007–9021 (2003).
Wierstra, I. & Alves, J. Cyclin E/Cdk2, P/CAF, and E1A regulate the transactivation of the c-myc promoter by FOXM1. Biochem. Biophys. Res. Commun. 368, 107–115 (2008).
Chen, H., Liu, H. D. & Qing, G. L. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 3, 5 (2018).
Caforio, M. et al. Recent advances in searching c-Myc transcriptional cofactors during tumorigenesis. J. Exp. Clin. Cancer Res. 37, 239 (2018).
Hann, S. R. MYC cofactors: molecular switches controlling diverse biological outcomes. Cold Spring Harb. Perspect. Med. https://doi.org/10.1101/cshperspect.a014399 (2014).
Mugrauer, G. & Ekblom, P. Contrasting expression patterns of 3 members of the Myc family of protooncogenes in the developing and adult mouse kidney. J. Cell Biol. 112, 13–25 (1991).
Caugant, D. A. et al. Genetic diversity in relation to serotype in Escherichia coli. Infect. Immun. 49, 407–413 (1985).
Leffler, H. & Svanborg Eden, C. Glycolipid receptors for uropathogenic Escherichia coli on human-erythrocytes and uroepithelial cells. Infect. Immun. 34, 920–929 (1981).
Dobrindt, U. et al. S-fimbria-encoding determinant sfa(I) is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69, 4248–4256 (2001).
Middendorf, B. et al. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 186, 3086–3096 (2004).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
The UniProt Consortium. UniProtKB – A0A454A9A9 (A0A454A9A9_ECOL5) https://www.uniprot.org/uniprot/A0A454A9A9 (2019).
Ramsay, G., Evan, G. I. & Bishop, J. M. The protein encoded by the human proto-oncogene c-Myc. Proc. Natl Acad. Sci. USA 81, 7742–7746 (1984).
Li, L. K. et al. GLIPR1 suppresses prostate cancer development through targeted oncoprotein destruction. Cancer Res. 71, 7694–7704 (2011).
Mashhoon, N. et al. Crystal structure of a conformation-selective casein kinase-1 inhibitor. J. Biol. Chem. 275, 20052–20060 (2000).
Uhlen, P. et al. alpha-Haemolysin of uropathogenic E. coli induces Ca2+ oscillations in renal epithelial cells. Nature 405, 694–697 (2000).
Patzer, S. I. & Hantke, K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28, 1199–1210 (1998).
Yatsunyk, L. A. et al. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 13, 271–288 (2008).
Cogswell, J. P. et al. Mechanism of C-Myc regulation by c-Myb in different cell lineages. Mol. Cell. Biol. 13, 2858–2869 (1993).
Nakagoshi, H., Kaneiishii, C., Sawazaki, T., Mizuguchi, G. & Ishii, S. Transcriptional activation of the c-Myc gene by the c-Myb and b-Myb gene-products. Oncogene 7, 1233–1240 (1992).
Balamurugan, K. & Sterneck, E. The many faces of C/EBPdelta and their relevance for inflammation and cancer. Int. J. Biol. Sci. 9, 917–933 (2013).
Gery, S., Tanosaki, S., Hofmann, W. K., Koppel, A. & Koeffler, H. P. C/EBPdelta expression in a BCR-ABL-positive cell line induces growth arrest and myeloid differentiation. Oncogene 24, 1589–1597 (2005).
Whitfield, J. R., Beaulieu, M. E. & Soucek, L. Strategies to inhibit Myc and their clinical applicability. Front. Cell Dev. Biol. 5, 10 (2017).
Mossberg, A. K., Hou, Y. C., Svensson, M., Holmqvist, B. & Svanborg, C. HAMLET treatment delays bladder cancer development. J. Urolog. 183, 1590–1597 (2010).
Tran, T. H. et al. Bladder cancer therapy without toxicity—a dose‐escalation study of alpha1‐oleate. Int. J. Cancer 147, 2479–2492 (2020).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 65, 5–29 (2015).
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J .Clin. 65, 87–108 (2015).
Vos, T. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet 390, 1211–1259 (2017).
Ren, J., Sui, H., Fang, F., Li, Q. & Li, B. The application of ApcMin/+ mouse model in colorectal tumor researches. J. Cancer Res. Clin. Oncol. 145, 1111–1122 (2019).
Ambite, I. et al. Fimbriae reprogram host gene expression – divergent effects of P and type 1 fimbriae. PLoS Pathog. 15, e1007671 (2019).
Lutay, N. et al. Bacterial control of host gene expression through RNA polymerase II. J. Clin. Invest. 123, 2366–2379 (2013).
Lee, I. & Suzuki, C. K. Functional mechanics of the ATP-dependent Lon protease—lessons from endogenous protein and synthetic peptide substrates. Biochim. Biophys. Acta Proteins Proteom. 1784, 727–735 (2008).
Pelengaris, S., Khan, M. & Evan, G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2, 764–776 (2002).
Soucek, L. et al. Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 (2008).
Zdziarski, J. et al. Host imprints on bacterial genomes-rapid, divergent evolution in individual patients. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1001078 (2010).
Ambite, I. et al. Active bacterial modification of the host environment through RNA polymerase II inhibition. J. Clin. Invest. https://doi.org/10.1172/JCI140333 (2020).
Oeyen, S. et al. Long-term outcome after acute kidney injury in critically-ill patients. Acta Clin. Belg. 62, 337–340 (2014).
Leek, J. T. & Storey, J. D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, 1724–1735 (2007).
Mobley, H. L. T. et al. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells – role of hemolysin in some strains. Infect. Immun. 58, 1281–1289 (1990).
Eisenhart, C. The assumptions underlying the analysis of variance. Biometrics 3, 1–21 (1947).
Acknowledgements
We thank the Swedish Research Council, Swedish Cancer Society, H.J. Forssman Foundations, Medical Faculty (Lund University), Royal Physiographic Society, Network of Excellence: Infect-ERA and ALF grants from the Medical Faculty and Regional Laboratories (Labmedicin Skåne). The work of the Münster group was supported by the German Ministry of Education and Research (no. 031L0007B) and the German Research Foundation (nos. SFB 1009 and TP B05). We also thank the Swedish National Infrastructure for Biological Mass Spectrometry. Support to the Svanborg group was further provided from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 954360.
Author information
Authors and Affiliations
Contributions
D.S.C.B., C.C., J.P., M.L.Y.W., T.H.T., I.A., U.D. and C.S. designed the study, performed the experiments, analyzed the data and wrote the paper. D.S.C.B., C.C., M.L.Y.W. and S.A. performed in vitro experiments. T.H.T. performed animal experiments and immunohistochemistry. S.M.C. collected clinical samples. C.C., I.A. and M.L.Y.W. carried out bioinformatics. U.D. and J.P. constructed bacterial mutants. S.K. and C.W. performed mass spectrometry analysis. Statistics were analyzed by D.S.C.B. and M.L.Y.W.
Corresponding author
Ethics declarations
Competing interests
A patent has been filed to protect the use of bacterial MYC inhibition therapy (inventors D.S.C.B., C.C. and C.S.).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Control experiments for Main Figs. 1 and 2.
a, b, Effects of infection on cell viability. Human kidney cells were infected with E. coli 83972 (ABU), CFT073 (APN) or 536 (APN) (105 CFU/ml, 4 hours). PBS served as a control. Cell viability was measured by (a) Lactate dehydrogenase release (n = 3 experiments) and (b) Presto Blue levels (n = 2 experiments). c, Low magnification confocal images of human kidney cells infected with different E. coli 536 deletion mutants (PAI I536 to PAI V536 and PAI I-II536) (105 CFU/ml, 4 hours). Cellular c-MYC staining visualised by confocal microscopy (green = c-MYC, blue = nuclei, scale bar = 10 μm) (left panel). Quantification of fluorescence intensity in 50 cells (n = 3 experiments) (right panel). Data are presented as mean ± s.d. and analysed by two-tailed paired t-test. ** P < 0.01, *** P < 0.001. d, Growth curves of bacterial strains used in this study. A single colony of each E. coli strain or mutant was cultured in Luria Broth in a shaking incubator at 37 °C. OD600 was measured at 1, 2, 3, 4, 6 and 8 hours. Average of n = 2 experiments per strain.
Extended Data Fig. 2 Control experiments for Main Fig. 3.
a, Comparative analysis of bacterial proteins in supernatants of E. coli 536 and PAI I536. Five bands, abundant in E. coli 536 supernatant (SN) (bands 1-5, Coomassie blue staining), were excised, processed by in-gel digestion and analysed by LC-MS/MS. b, The 10 most abundant proteins from each excised band are listed. c, c-MYC peptide fragments detected by LC-MS/MS. 23 specific peptides corresponding to 51% of the c-MYC sequence were detected in the control sample. Ten specific peptides corresponding to 23% of the sequence were found after recombinant Lon protease (rLon) degradation of c-MYC. d, Binding of rLon protease to recombinant c-MYC in a dose-dependent manner (n = 2 experiments). e, Protease digestion of E. coli 536 SN removed MYC inhibition, suggesting that the soluble bacterial MYC inhibitor is a protein. Supernatants were pre-treated with increasing concentrations of trypsin (0-5 μg/ml, 30 min) before being added to human kidney cells (4 hours). Trypsin was used as a negative control (n = 2 experiments). f, g, Human kidney cells were treated with inhibitors of ATPase or GTPase driven endocytosis (Quercetin [25 μM] or Dynasore [30 μM]) for 30 minutes prior to rLon protease treatment (8 μg/ml, 4 hours). Confocal images show reduced Lon uptake (red = Lon, blue = nuclei, scale bar = 10 μm) (f). Quantification of Lon uptake in 50 cells (n = 3 experiments) (g). Data are presented as mean ± s.d. and analysed by two-tailed paired t-test. * P < 0.05. N.S. = No significance.
Extended Data Fig. 3 Control experiments for Main Fig. 4.
a, CK1α1-dependent c-MYC degradation. Genes involved in proteasome degradation were upregulated in E. coli 536 infected human kidney cells, including CSNK1A1 and PLK3 (Red = up-regulated, black = not regulated, cut off FC > 1.41). b, CSNK1A1 expression was increased by E. coli 536 infection and by the E. coli 536 supernatant (SN) exposure (n = 4 experiments). c, CK1α1 activation, defined by a redistribution from the nuclei to characteristic cytoplasmic puncta (green = CK1α1, blue = nuclei, n = 3 experiments, scale bar = 10 μm). d, Binding of CK1α1 to c-MYC shown by co-immunoprecipitation, using recombinant c-MYC and lysates from infected cells (n = 3 experiments). e, Reversal of c-MYC inhibition by the CK1α1 inhibitor IC261 (30 min pre-treatment) by confocal imaging (green = c-MYC, blue = nuclei, n = 5 experiments, scale bars = 10 μm). Quantification of fluorescence intensity in 50 cells (black = PBS, red = E. coli 536). f, Western blot analysis (n = 4 experiments). g, Ca 2+ fluxes in human kidney cells were inhibited by EGTA treatment (50 μg/ml, 30 min) prior to stimulation with supernatants from E. coli 536 WT or 536 Δhly1/2. EGTA pre-treatment inhibited the activation of CK1α1. Data are represented as mean ± s.d. and analysed by two-tailed one-way ANOVA with Geisser greenhouse correction (b) or two tailed paired t-test (e). * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 4 Role of hly I and hly II for MYC inhibition.
a, Inhibition of c-MYC staining in human kidney cells infected with E. coli 536 or exposed to its supernatant (SN). Partial inhibition by E. coli 536 Δhly1/2 or its supernatant. b, Quantification of fluorescence intensity in (a). (50 cells, n = 2 experiments). c, Western blot analysis of the corresponding cell extracts. d-f, Molecular size filtration of the E. coli 536 SN. The α-hemolysin (HlyA) protein is 109 kDa, predicting that HlyA should be excluded from the < 100 kDa fraction. A second fraction of < 30 kDa was also collected. The <100 kDa fraction inhibited c-MYC as shown by confocal imaging (n = 3 experiments, scale bars = 10 μm) in (d) and quantification in 50 cells in (e), and Western blots analysis (n = 3 experiments) in (f). The < 30 kDa fraction was less active. The < 100 kDa fraction did not affect cell viability quantified by ATP lite (compared to untreated cells, n = 3 experiments). h, Hemolysin (Hly)- and Lon protease (Lon) production in clinical APN and ABU isolates from the collection shown in Fig. 1c. Hemolysin (Hly) production was detected on blood agar. h, c-MYC degradation was most efficient for APN strains that expressed both Lon and Hly (Western blot analysis). Data are presented as mean ± s.d. and analysed by two-tailed paired t-test. *P < 0.05, ** P < 0.01, *** P < 0.001 (e, g) and Mann-Whitney U-test (h). N.S. = no significance.
Extended Data Fig. 5 Control experiments for Main Fig. 4.
a, Effects of hly1/2 and lon deletions on transcriptional MYC regulators MAX, c-MYB and C/EBP-δ. Human kidney cells were infected with E. coli 536, 536 Δhly1/2 or 536 Δlon (105 CFU/ml, 4 hours). a, d, g, MAX levels were not affected. b, e, h, c-MYB levels were reduced by E. coli 536 and Δlon but not mRNA levels. c, f, i, C/EBP-δ levels were affected by E. coli 536. Confocal images (scale bar = 10 μm) in (a). d-f Quantification of images in (a-c) (50 cells per image, n = 3 experiments). MAX (d), c-MYB (e) and C/EBP-δ (f) staining intensity compared to PBS. g, MAX, h, MYB and i, CEBPD mRNA levels quantified by qRT-PCR. PBS served as control (n = 2 experiments). Data are presented as mean ± s.d. and analysed by two-tailed one-way ANOVA with Geisser-Greenhouse correction (d-i). * P < 0.05, ** P < 0.01, *** P < 0.001. j, k, Cell viability as quantified by LDH release after in vitro infection of human kidney cells. j, No change in cell viability after exposure to bacterial supernatants quantified by LDH release (n = 3 experiments). k, No change in viability of cells exposed to increasing concentrations of recombinant Lon protease (rLon), quantified by LDH, ATP lite or Presto Blue (n = 2 experiments). N.S. = No significance.
Extended Data Fig. 6 Control experiment for Main Fig. 5.
a, c-MYC inhibition in different tumour cell lines (HTB-9, MB49, A549, DLD1) following bacterial infection (for supernatant data, see main Fig. 5b). The effect of E. coli 536 was compared to E. coli PAI I536, 536 Δhly1/2 or 536 Δlon mutants (105 CFU/ml, 4 hours). Western blot of c-MYC levels (n = 3 experiments per cell line). b-f, Therapeutic effect of E. coli 536 supernatant (SN) in a bladder cancer model. E. coli PAI I536 SN showed no therapeutic activity. Scale bar = 2 mm. b, Tumour development was delayed on day 12 in mice receiving E. coli 536 SN but not E. coli PAI I536 SN. c, Tumour size was reduced, as quantified in whole bladder tissue mounts. Representative images (n = 5 per group). d, c-MYC expression was reduced in mice treated with E. coli 536 SN compared to mice receiving PBS or PAI I536 SN. e, Western blot of whole bladder tissue extracts (n = 5 per group). f, Transcriptomic analysis of whole bladder RNA. The shift from a disease-specific towards a healthy tissue profile in E. coli 536 treated mice was not seen in the E. coli PAI I536 group (Principle Component Analysis (PCA) plot). g, Quantification of bladder weight, bladder size, tumour size and c-MYC staining (n = 9 Sham and 536 SN, n = 5 PAI I536 SN). Data are presented as mean ± s.d. and analysed by Kruskal-Wallis test with Dunn’s Correction. Data from Sham and recombinant Lon protease (rLon) treated mice is also included in main Fig. 5.
Extended Data Fig. 7 Comparative gene expression analysis of bladder tissues 12 days.
a, Gene categories regulated by E. coli 536 supernatant (SN) compared to Sham. Predominance of cancer-related genes including bladder cancer genes. b, Cancer functions regulated in the E. coli 536 SN treated mice compared to Sham. The majority were down-regulated. c, Heat-map comparing E. coli 536 SN- or recombinant Lon protease (rLon)-treated mice to Sham (cut off FC ≥ 2, P < 0.05 compared to healthy bladders). d, Principle Component Analysis (PCA) plot showing a shift from disease (Sham) to a healthier phenotype (Healthy), which was more pronounced in rLon treated than in E. coli 536 SN treated mice. e, A similar trend was seen by the biofunctions analysis, illustrating the comparison of rLon treated mice and E. coli 536 SN treated mice. Data from rLon treated mice compared to Sham are also included in main Fig. 5.
Extended Data Fig. 8 Long-term effects of recombinant Lon protease treatment in the MB49 bladder cancer model.
a-d, Prolonged protection was detected in recombinant Lon protease (rLon) treated mice at follow-up without further treatment (27 days) and with further treatment of rLon twice per week (35 days) compared to Sham, which were sacrificed on day 12 (n = 15 Sham and rLon follow-up, n = 12 rLon long-term, n = 2 experiments). a, A decrease in pathology score was detected in rLon treated mice after 27 days and after 35 days. b-d, Lower bladder weight in rLon treated mice at follow-up (27 days) and after continued treatment (35 days) compared to Sham (day 12). Smaller bladder size in mice receiving continued treatment compared to Sham (day 12). e, Heat-map showing a lower number of regulated genes in the follow-up and long-term groups compared to Sham (red = up-regulated, blue = down-regulated, black = not significantly regulated, cut off FC ≥ 2, P < 0.05, n = 2 per group). Data are presented as mean ± s.d. and analysed by two-tailed Kruskal-Wallis test with Dunn’s correction. f, Top regulated genes associated with the Molecular Mechanisms of Cancer pathways were strongly inhibited by rLon treatment at follow-up and further in the long-term treatment group.
Extended Data Fig. 9 Long-term effects of recombinant Lon protease treatment in ApcMin/+ mice.
a, Gene expression analysis comparing recombinant Lon proteae (rLon) to Sham after long-term follow-up (W25). Pie chart of regulated gene categories. rLon treatment mainly affected cancer related gene functions, including those associated with colon cancer. b, Details of cancer-related functions regulated in rLon treated mice compared to Sham. c, Network showing marked inhibition of MYC-related genes (red = up-regulated, blue = down-regulated). MYC inhibition was maintained in the long-term follow-up group.
Extended Data Fig. 10 Control experiment for Figs. 5 and 6.
a-d, Analysis of recombinant Lon protease (rLon) toxicity in healthy mice. a, b, C57BL/6 J WT mice were exposed to rLon intra-vesically for 12 days. No change in bladder weight or macroscopic appearance of the bladder was detected (n = 5 mice per group). Scale bar = 2 mm. c, Few genes were regulated in bladders from the rLon treated mice (n = 2 rLon, n = 1 Healthy control). d, MYC expression was not regulated but a small number of MYC-dependent genes was regulated in bladders from rLon treated healthy mice (32 activated, 1 inhibited). e-h, Analysis of per-oral rLon protease challenge in healthy C57BL/6 J WT mice, exposed to rLon per-orally twice daily for 14 days. f, No change in body weight, or macroscopic appearance of the intestine, liver, kidney or spleen was detected (n = 3 mice per group). Scale bar = 5 mm. g, Gene expression analysis detected a low number of regulated genes in intestinal segments of rLon treated mice (n = 3 rLon, n = 1 Healthy). h, MYC expression was not regulated and few MYC-related genes were affected after rLon challenge (1 activated, 8 inhibited genes) (FC compared to unchallenged controls, cut off FC ≥ 2, P < 0.05). Data are presented as mean ± s.d., N.S. = No significance.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1–4.
Source data
Source Data Fig. 1
Unprocessed immunoblots for Fig. 1.
Source Data Fig. 2
Unprocessed immunoblots for Fig. 2
Source Data Fig. 3
Unprocessed immunoblots for Fig. 3.
Source Data Fig. 4
Unprocessed immunoblots for Fig. 5.
Source Data Fig. 5
LS–MS/MS data for in vitro degradation.
Source Data Extended Data Fig. 2
Unprocessed gels: immunoblots for Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Unprocessed immunoblots for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Unprocessed immunoblots for Extended Data Fig. 4.
Source Data Extended Data Fig. 6
Unprocessed immunoblots for Extended Data Fig. 6.
Rights and permissions
About this article
Cite this article
Butler, D.S.C., Cafaro, C., Putze, J. et al. A bacterial protease depletes c-MYC and increases survival in mouse models of bladder and colon cancer. Nat Biotechnol 39, 754–764 (2021). https://doi.org/10.1038/s41587-020-00805-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-00805-3
This article is cited by
-
Modified screening of MYC promotor region elements using the CRISPR library in ovarian cancer
Journal of Ovarian Research (2025)
-
Multitarget mechanism of MYC inhibition by the bacterial lon protease in disease
Scientific Reports (2025)
-
MYC in cancer: from undruggable target to clinical trials
Nature Reviews Drug Discovery (2025)
-
Comprehensive retrospect and future perspective on bacteriophage and cancer
Virology Journal (2024)
-
Bioactive nutraceuticals as G4 stabilizers: potential cancer prevention and therapy—a critical review
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)